ASTRA’s new LNP-RNA Analysis quantifies payload,
encapsulation efficiency and more
Novel modalities require extensive characterization
mRNA rose to fame as a new and effective therapeutic modality through its extraordinary success as a COVID vaccine. However, these mRNA-based vaccines would not exist without formulation by lipid nanoparticles (LNPs). The lipid shell protects mRNA from nuclease degradation and endosomal digestion when it enters the cell. As a result, LNP is becoming the preferred delivery vehicle for different types of RNA- and DNA-based medicines.
Since LNP formulation for medicinal nucleic acids are novel and complex, the pharmaceutical industry is still learning how to properly manufacture and characterize LNPs loaded with RNA or DNA. Essential attributes that must be analyzed to ensure safe and efficacious drug products include:
- nanoparticle size distribution
- particle concentration
- overall lipid and nucleic acid concentration
- particle morphology
- stability under applied stresses including freeze-thaw, presence of bio-medium, and elevated temperatures
However, LNPs must also be characterized for more complex attributes such as encapsulation efficiency and the amount of encapsulated nucleic acid. Since transduction of the genetic material into the cell depends in large measure on nanoparticle size, and LNPs tend to be rather heterogeneous in size, determining the genetic payload for each nanoparticle size fraction is to some degree a ‘holy grail’: together with overall titer, the size-based payload distribution is invaluable in predicting and controlling nanopharmaceutical efficacy.
Numerous and sometimes difficult analytical methods
The graphic below shows some major quality attributes of LNP-RNA and, in the green rectangles, the traditional techniques used to quantify them.
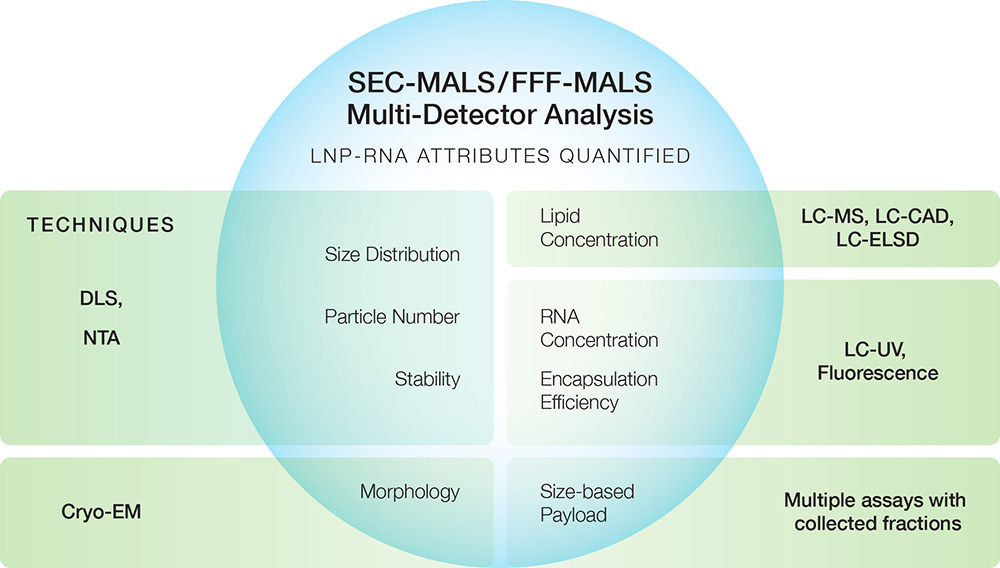
Many of these techniques for quantifying LNP attributes have significant limitations in resolution, precision, accuracy and speed. For example, dynamic light scattering (DLS) is the primary tool to quantify size, polydispersity and stability of the loaded LNP samples. Though DLS is a popular and useful screening tool, its limited resolution prevents it from detecting small yet potentially important differences between processes and batches. In addition, DLS does not reveal payload information or structural details.
Another such example is related to payload quantitation. Specifically, for RNA quantitation, fluorescence measurement with RiboGreen dye is a popular assay. The accuracy of this assay is, however, easily compromised by the presence of several commonly used salts and the interaction between the dye and the PEGylated lipid. Ion-exchange chromatography (IEC) with UV detection is thus used as an orthogonal assay to confirm RNA concentration. Even after the assay to measure RNA concentration is identified and validated, it is still tedious and time-consuming to profile RNA payload as a function of size – an attribute that helps researchers understand the true efficacy of the therapy. To measure the size-dependent RNA payload, the current method calls for collecting fractions from an LNP size separation system, SEC or more often FFF, measuring the LNP size by DLS, then rupturing the lipid particles using a surfactant, and finally using fluorescence or IEC-UV to quantify RNA. Because it is difficult to measure RNA payload as a function of size, such data are scarce in the literature.

The Wyatt solution: one technique to measure them all
As the workhorses for biopharmaceutical and bionanoparticle characterization, SEC-MALS and FFF-MALS had also been adopted as extended characterization tools for LNP-encapsulated RNA or DNA. The data from online MALS, UV, and dRI detectors following SEC or FFF are used for routine yet in-depth analyses, such as high-resolution size distributions, particle concentration and morphology in a single run. This versatility enables SEC- and FFF-MALS to excel in stability and lot-to-lot studies, thanks to their ability to identify and highlight even small changes to the samples.
However, in attempting to quantify the contents of the LNP-RNA entity, these methods ran into an obstacle. As with the analysis of smaller conjugates such as glycoproteins or adeno-associated virus, two concentration sources are needed to determine the amount of lipids and the amount of nucleic acids in each nanoparticle: refractive index (RI) and UV absorbance. With sufficient sample concentration, RI signals can be measured readily and accurately. However, the UV signals are inflated due to scattering of the UV light in the detector by the particles. In fact, a nanoparticle made of purely non-absorbing lipids still exhibits a significant UV signal due to this scattering phenomenon. This is generally true for particles with radii above ~ 20 – 25 nm, and the degree of UV scattering increases relative to the degree of UV absorption with increasing particle size.
Recently, scientists at Wyatt Technology have developed a new method that uses data from the online MALS, UV and RI detectors to quantify LNP payload. The method removes the scattering contribution from the UV signal of the nanoparticle, and then applies a calculation similar to standard conjugate analysis. When combined with size-based separation (SEC or FFF), the new analysis hence enables quantitation of RNA molar mass and concentration as a function of size with no need to analyze fractions off-line. This novel LNP payload analysis method (US patent pending) has now been fully implemented in the LNP Analysis module of ASTRA 8.1.
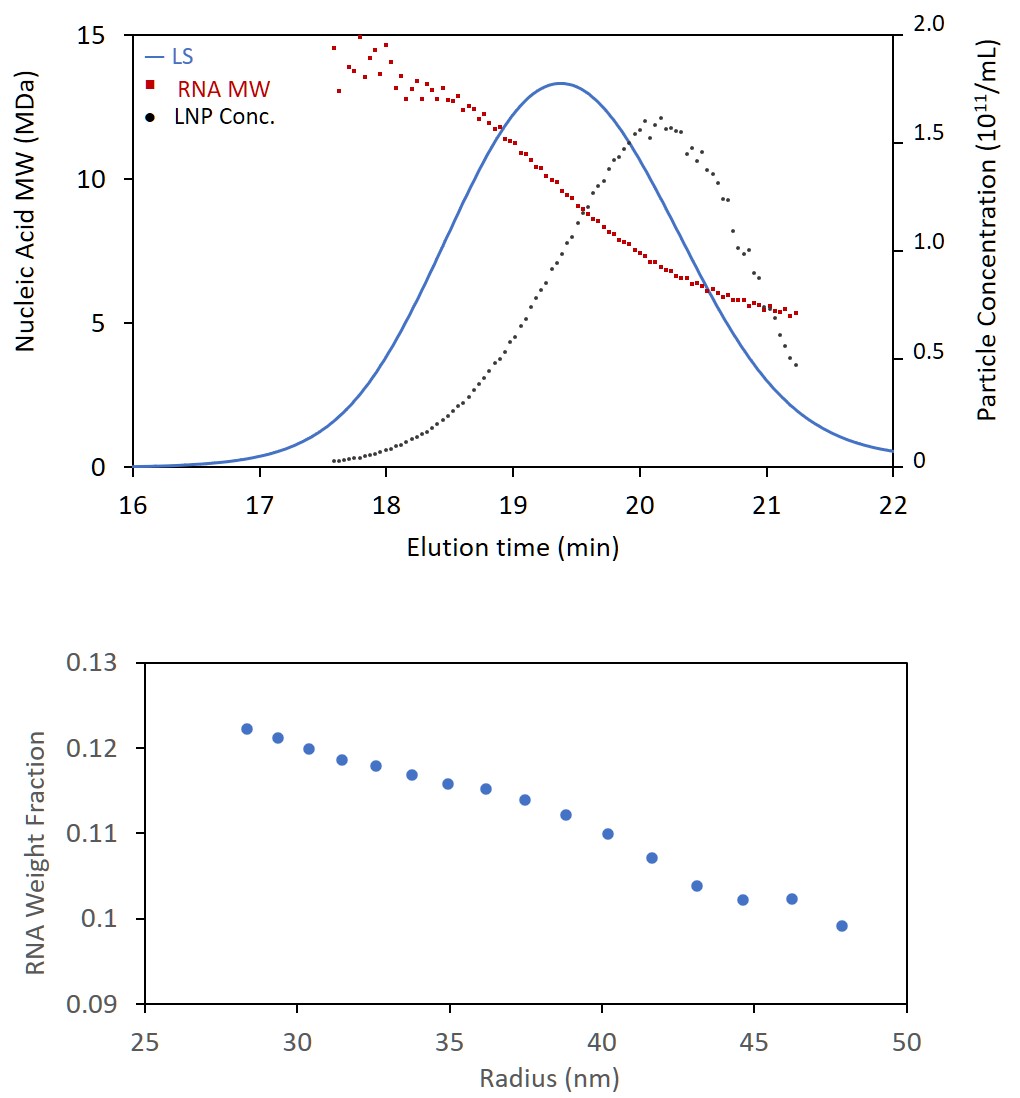
Comprehensive and accurate characterization in a single run facilitates safe and efficacious therapeutics
With the ability to determine the nucleic acid content across the chromatogram or fractogram, it is now possible to calculate encapsulation efficiency, either by comparing the total genetic content of the LNPs to the amount added during formulation, or to the amount of free nucleic acid which elutes as a separate peak. With the new analysis, most, if not all, of the attributes in the above graphic can be characterized and quantified by a single run on one system. What’s more, the method requires no special sample prep or reagents like RiboGreen.
To cross-verify the online LNP analysis method for payload distribution and encapsulation efficiency, we have worked with a number of LNP research groups using the traditional method for measuring size-dependent RNA payloads. One such study, led by Dr. Xiujuan Jia and her colleagues at Merck, was recently published (https://authors.elsevier.com/c/1e0rr5a~s4O8gd). Echoing what Dr. Jia emphasized in her presentation at the 2021 Eastern Analytical Symposium – “Novel delivery requires a new way of thinking” – we believe that the combination of our SEC-MALS and FFF-MALS systems with ASTRA’s new LNP Payload Analysis enables researchers to gain a deeper understanding of their novel delivery vehicle, making RNA and DNA therapeutics safer, more consistent and effective.

