Is it possible to determine molar mass, size, and relative fractions of a copolymer or protein conjugate with light scattering?

Introduction
When combining your light scattering detector with an RI and UV detector, you can utilize ASTRA™ to determine the constituent fractions. This is especially helpful for determining the fraction and molar mass of components in a number of two-component systems such as protein-nucleic acid complexes, PEGylated proteins, glycosylated proteins, membrane proteins associated with surfactants. It can also be applied to copolymers where the components have distinct UV absorption coefficients.
ASTRA has native support for this type of analysis in the Protein Conjugate Analysis procedure, which can be added or applied to any experiment with triple detection (LS, UV, RI).
System validation
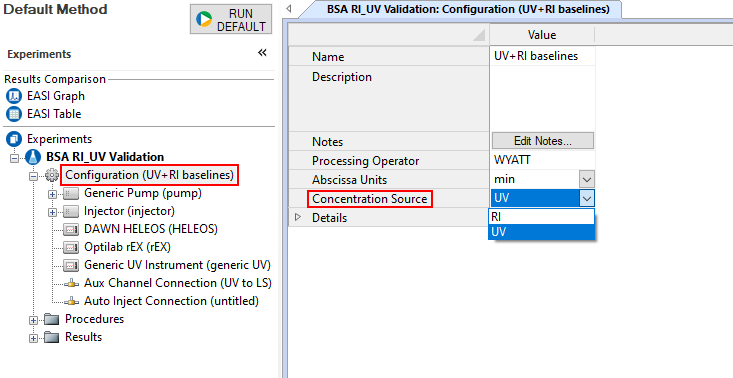
Before using the Protein Conjugate Analysis method, it is important to validate your system on a non-conjugated protein or polymer to ensure your RI and UV signals give accurate molar mass data. This can be done with a standard such as BSA, where we would expect the monomer molar mass from both the RI and UV detectors to be the same. You can toggle between the RI and UV as the concentration source in the Configuration Window of your experiment to confirm that each source calculates the correct molar mass.
Applying the protein conjugate analysis method
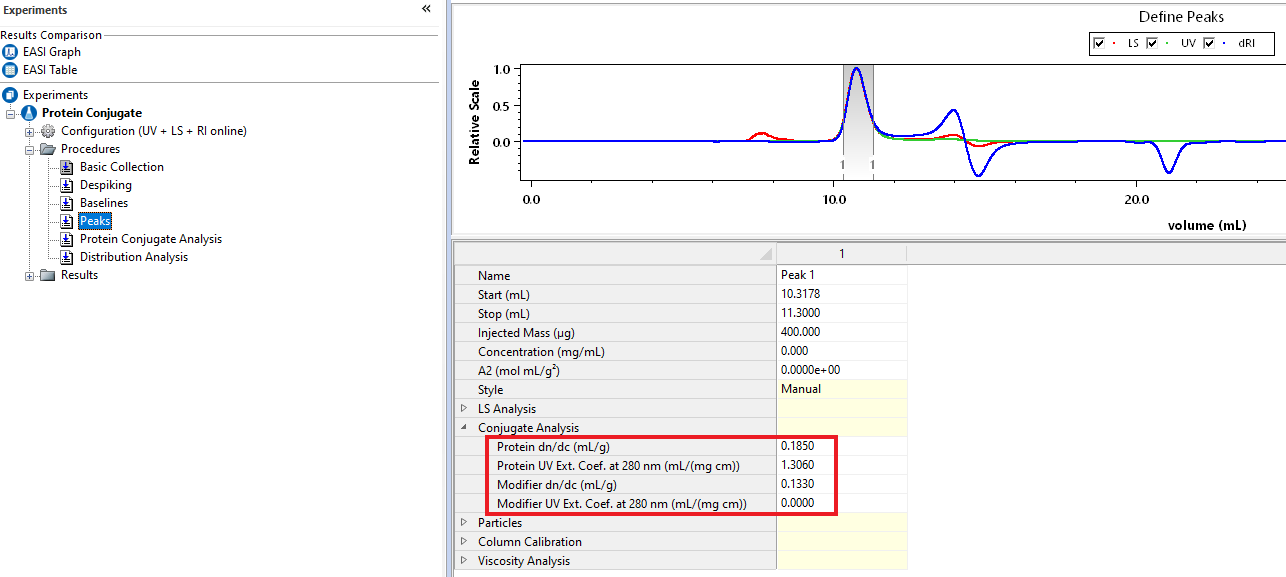
To differentiate between the first component, termed the ‘protein,’ and the second component, referred to as the ‘modifier,’ you’ll need to know the dn/dc and UV extinction coefficients for each component. (For a copolymer, these would be the values for your two different polymers). These constants can be found in the literature or can be directly measured using either batch or online measurements with ASTRA. Once you have the necessary constants, enter them in ASTRA under the Peaks Procedure.
Using this information with the three detectors, ASTRA can compute the following information:
- Molar mass of the entire complex
- Molar mass of the protein portion of the complex
- Molar mass of the modifier (non-protein) portion of the complex
- Mass fraction of the modifier and protein
To get this information, apply the Protein Conjugate Method located in ASTRA to your existing data file. You can do this by right-clicking your experiment file and clicking Apply Method... and navigating to System -> Methods -> Light Scattering -> Protein Conjugate. From each slice of data, ASTRA will compute the overall molar mass for your peak.
Protein conjugate analysis in EASI Graph
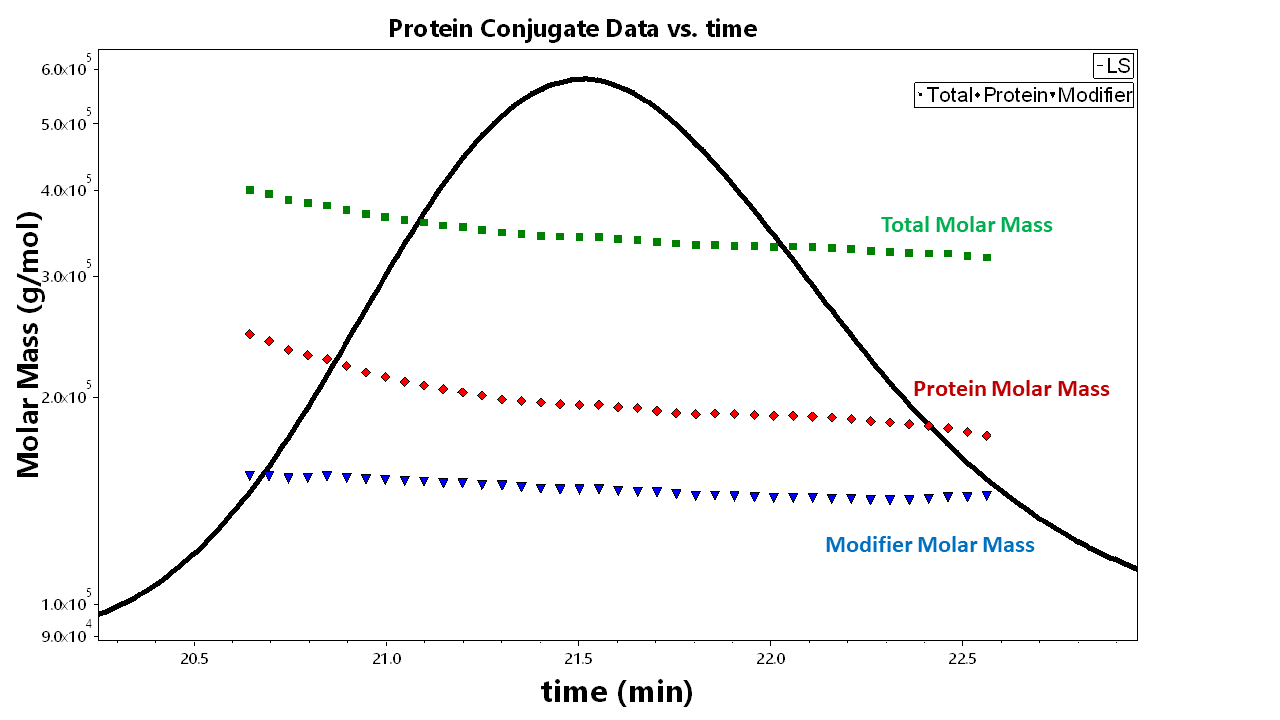
You can view this information in EASI Table or the ASTRA report and view a graph of the protein conjugate analysis in EASI Graph (you can do this by selecting “Protein conjugate” as the display option). EASI Table can report the protein and modifier molar mass data (as well as other information based on the scalars you select).
The EASI Graph plot on the right shows the result of the protein conjugate analysis performed on a membrane protein with surfactants. This plot shows the total molar mass over the entire peak selection with the respective molar masses of the protein and surfactants.
Conclusion
You can find additional information and tutorials about the Protein Conjugate Analysis and the graphing features available in ASTRA User’s Guide and on the Support Center, including step-by-step instructions in TN1006: Performing Protein Conjugate Analysis.
Do you have a question? Contact our experts here in Customer Support. We’re happy to help! Call +1 (805) 681-9009 option 4.