Conjugation & Payload Analysis
The solution-based analysis of macromolecular conjugates is vastly simplified by light scattering.
Introduction
Many interesting and useful macromolecules or nanoparticles consist of two chemically and physically distinct species, conjugated covalently or one encapsulating the other. This can make it quite difficult to characterize their essential properties.
Multi-angle light scattering (MALS) using a DAWN™ MALS instrument combined with UV absorption, an Optilab™ differential refractive index (dRI) detector and a separation technique are invaluable in analyzing conjugation ratio or payload, as well as size distributions, encapsulation efficiency, shape, conformation or polymer branching. Determining the quantities of each component in the whole and how these depend on size may be especially critical for drug products, vaccines and gene therapies.
Conjugated Macromolecules
Traditional analytical size-exclusion chromatography (SEC) is insufficient to characterize conjugates since no calibration standards exist to represent their elution behavior. The combination of MALS, UV and dRI detection with SEC, plus ASTRA's Conjugate Analysis algorithm, offers the solution to the characterization of macromolecules such as:
- Glycoproteins, lipoproteins and nanodiscs
- Protein-polysaccharide vaccines
- Block co-polymers
- PEGylated proteins
- Protein-nucleic acid complexes
- Membrane proteins solubilized in detergent micelles
How it works
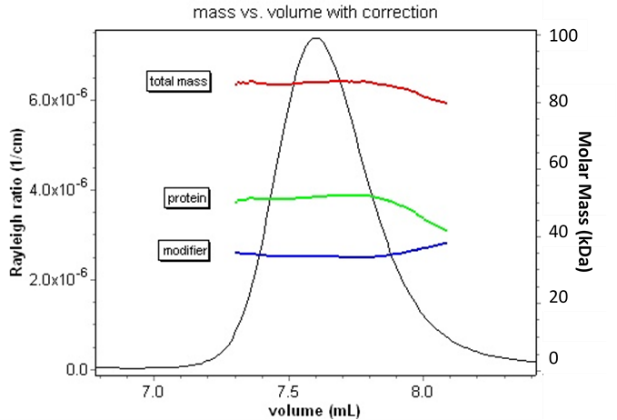
Conceptually, a complex consisting of a protein (containing a UV chromophore) and a modifier such as poly-ethylene glycol, or PEG (which does not), is analyzed by combining information from two distinct measurements. UV absorption determines the protein concentration, while dRI determines total macromolecular concentration. These two measurements are sufficient to determine the ratio of the protein and modifier in the complex (this is true even if both components absorb UV, as long as their absorbances differ sufficiently).
However, this analysis cannot determine the total mass or oligomeric state of the species. The addition of MALS provides the answer to full conjugate characterization. Since light scattering is proportional to the product of molar mass and concentration, the combination of these three signals is sufficient to determine not only the ratio but the actual molar mass, and hence oligomeric state, of each constituent. This information is critical in assessing aggregation of glycoproteins, determining the molecular conformation of conjugated polysaccharides, measuring the molar mass of block co-polymers, or deciding whether or not a detergent-solubilized membrane protein is present as a native oligomer.
AAV and small VLP
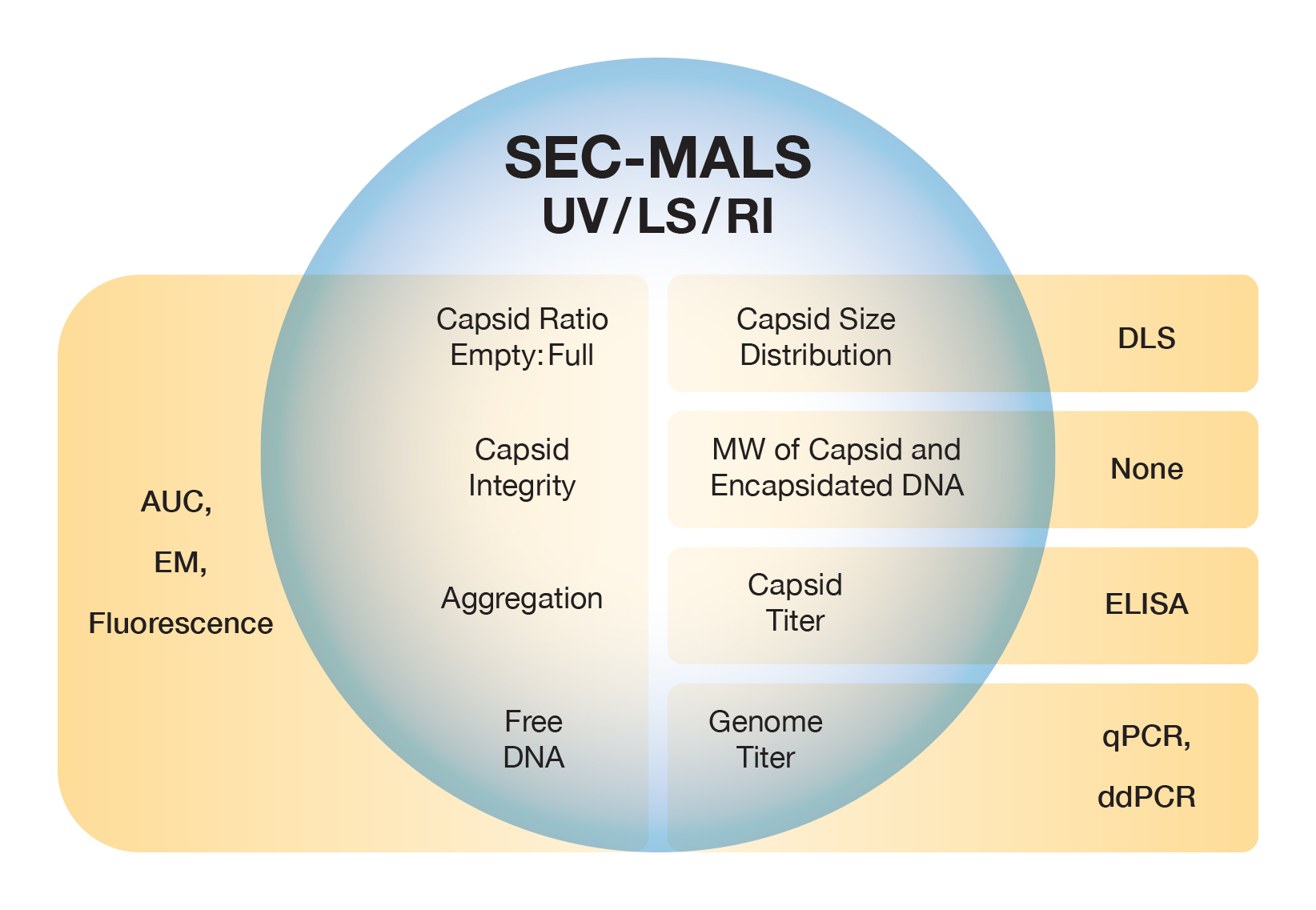
Adeno-associated virus and small, non-enveloped virus-like particles are special cases of the standard conjugate analysis. Their structural component tends to be very well defined in terms of the total molar mass of proteins that form the capsid, and the genomic payload is also quite specific. The SEC-MALS-UV-dRI method determines capsid molecular weight and the average payload molecular weight, from which the fraction of empty and full capsids may be calculated - even though they co-elute in SEC. Though less precise, the same analysis can be done with MALS and absorption at two UV wavelengths, usually 260 nm and 280 nm.
How it works
ASTRA’s Viral Vector Analysis extends the standard conjugate analysis to determine not just the molar mass of the protein and nucleic acid components, but also Vg/Cp, the ratio of full genomes to total capsids, and the overall concentration of total, empty and full capsids. It also quantifies aggregate levels, so in total three critical quality attributes (CQA) are quantified in a single run.
Since the SEC-MALS method is independent of AAV serotype, requires no reagents or special handling and is automated by standard HPLC instrumentation, it is suitable as a platform method covering multiple AAV products and all phases of development, manufacturing and QC. Wyatt offers extensive guidance on developing standard operating procedures for AAV analysis with SEC-MALS. Learn more about characterization of AAV vectors and Wyatt’s AAV Service Packages.
Nanoconjugates
Composite nanoparticles generally consist of a structural molecule and a payload. As for conjugated macromolecules, MALS-UV-dRI detection with a separation technique such as SEC or field-flow fractionation (FFF) is an essential method to characterize the size and composition of such binary composite nanoparticles. However, due to their larger size, there are certain challenges associated with analyzing nanoparticles by this procedure, which are overcome by ASTRA’s novel Nanoconjugate Analysis algorithm. Products that fall into this size range — and are amenable to nanoconjugate analysis ― include:
- Lipid nanoparticles (LNP) encapsulating RNA or DNA payloads
- Liposomes or PLGA polyplexes encapsulating small-molecule drug or peptide payloads
- Protein-polysaccharide vaccines in the multi-megadalton molar mass range
How it works
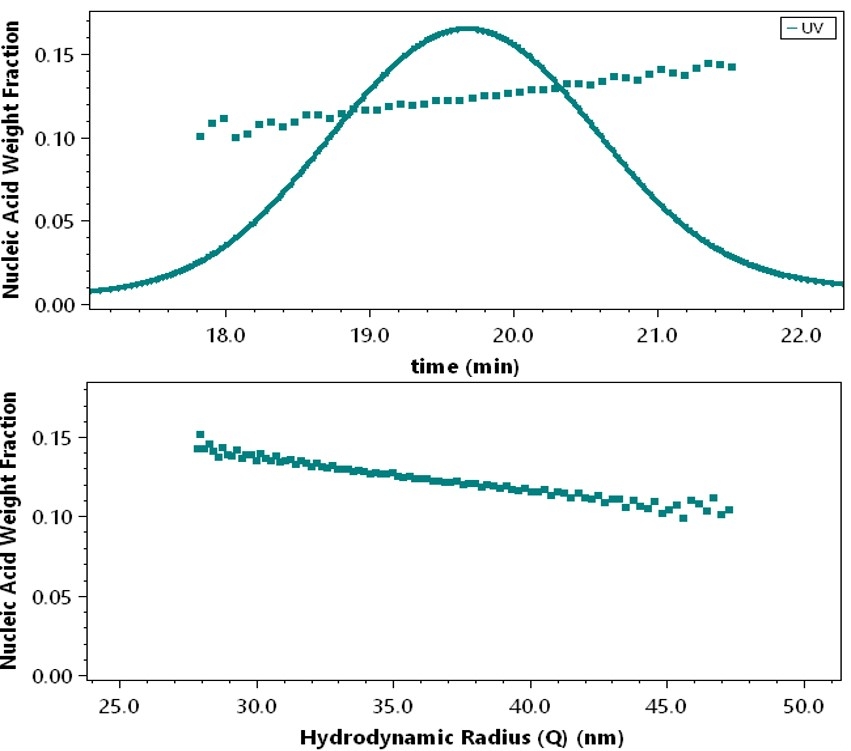
Conjugate analysis relies on accurate UV measurements. When the radius of the particle or large macromolecule is above roughly 30 nm, scattering of incident UV light in the detector by the analyte itself becomes significant. This results in a UV extinction signal reported by the detector, even though there might not be any absorption. For example, an empty liposome absorbs no UV at 260 nm but produces an appreciable UV signal due to scattering. Naively applying these UV data to the standard conjugate analysis produces erroneous results.
The Nanoconjugate Analysis characterizes the response of the UV detector to scattering by empty lipid particles or unconjugated polysaccharides over a size range that covers the sizes of the loaded nanoparticles or conjugated polymers. This information is then applied to a proprietary algorithm to quantify the nucleic acid or drug payload, or the conjugated protein, in the sample of interest. Since the analysis happens at each eluting slice, the payload is determined as a function of size.
Instrumentation
The Nanoconjugate Analysis may be used in conjunction with SEC if the samples are compatible with this method in terms of size and stability. Larger particles or those that may degrade under column shear may be separated by field-flow fractionation (FFF) using Wyatt’s Eclipse™ FFF system. The Eclipse is supported by industry-standard HPLC modules and combines seamlessly with Wyatt’s MALS and dRI detectors.
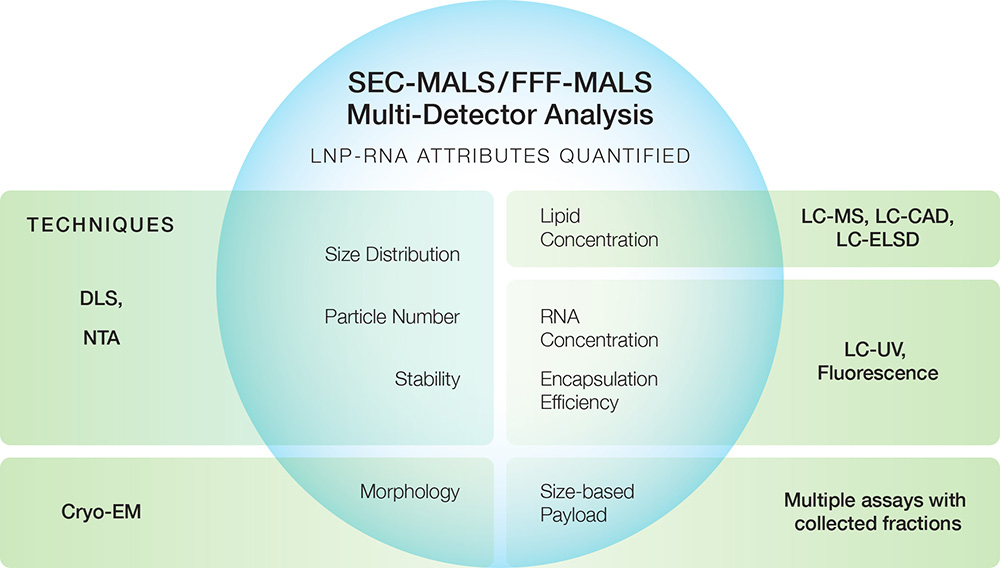
SEC-MALS or FFF-MALS with ASTRA’s LNP-RNA Analysis replace multiple traditional techniques for comprehensive biophysical analysis of LNPs encapsulating nucleic acids in a single run.
The LNP-RNA analysis provides the molecular weight of the nucleic acid payload at each eluting volume and as a function of size.
An Eclipse FFF-MALS system separates and characterizes macromolecules and particles with sizes 1 – 1000 nm.
Resources
Webinars
Virtual User Meeting – Protein Conjugate and Copolymer Analysis
Quantify Viral Vector Attributes with Light Scattering
Analyzing Ebola virus glycoprotein and its interactions with therapeutic antibodies using CG-MALS
Characterizing Protein Conjugates and Their Aggregates by Light Scattering
Application Notes
References
Abbas, S.; Lodge, T. P. Depletion interactions: effects of added homopolymer on ordered phases formed by spherical block copolymer micelles. Macromolecules 2008, 41, 8895-8902.
Bensaid, F.; du Boullay, O. T.; Amgoune, A.; Pradel, C.; Reddy, L. H.; Didier, E.; Sablé, S.; Louit, G.; Bazile, D.; Bourissou, D. Y-Shaped mPEG-PLA cabazitaxel conjugates: Well-controlled synthesis by organocatalytic approach and self-assembly into interface drug-loaded core-corona nanoparticles. Biomacromolecules 2013, 14, 1189-1198.
Berguig, G. Y.; Convertine, A. J.; Shi, J.; Palanca-Wessels, M. C.; Duvall, C. L.; Pun, S. H., Press, O. W.; Stayton, P. S. Intracellular delivery and trafficking dynamics of a lymphoma-targeting antibody-polymer conjugate. Mol. Pharm. 2012, 9, 3506-3514.
Citkowicz, A. et al. Characterization of virus-like particle assembly for DNA delivery using asymmetrical-flow field-flow fractionation and light scattering. Analytical Biochemistry 376,(2) 163-172 (2008).
McIntosh, N.L. et al. Comprehensive characterization and quantification of adeno-associated vectors by size exclusion chromatography and multi angle light scattering. Scientific Reports 11: 3012, (2021)
Peng, Y.; Zhang, L. Characterization of a polysaccharide-protein complex from Ganoderma tsugae mycelium by size-exclusion chromatography combined with laser light scattering. J. Biochem. Bioph. Meth. 2003, 56, 243-252.
Slotboom, D. J.; Duurkens, R. H.; Olieman, K.; Erkens, G. B. Static light scattering to characterize membrane proteins in detergent solution. Methods 2008, 46, 73-82.
Werle, A.K. et al., Comparison of analytical techniques to quantitate the capsid content of adeno-associated viral vectors. Molecular Therapy: Methods & Clinical Development 23, (December 2021)