Interactions
Biomolecular interactions are key to understanding a host of phenomena ranging from structure-function relationships and biotherapeutic activity through stability and aggregation.
Introduction
Whenever macromolecules change their behavior or properties as a consequence of proximity to another macromolecule, interactions are at play.
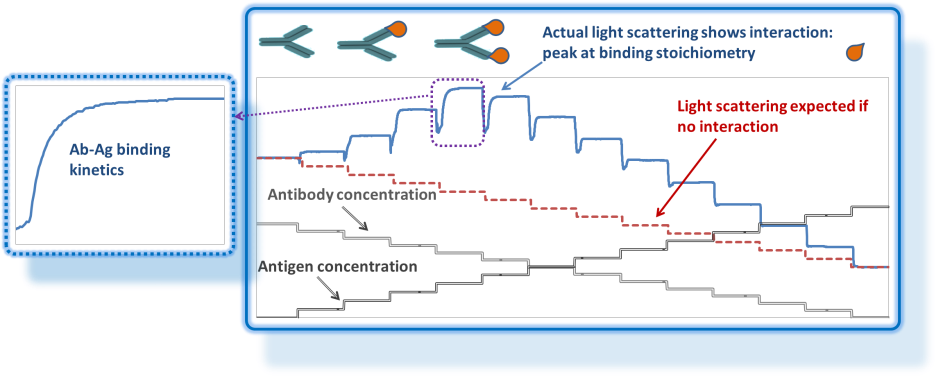
Reversible complexes are formed as a result of non-covalent interactions producing dynamic equilibrium between complexes and constituent monomers. This equilibrium is concentration-dependent, so a fractionation technique such as SEC-MALS - with the accompanying time-dependent dilution - is not suitable for full and rigorous characterization of reversible complexes. Among the most useful techniques for studying reversible complexes is composition-gradient light scattering (CG-MALS).
CG-MALS characterizes self- and hetero-association.
Non-covalent interactions generally fall into two classes:
- Specific interactions – those that lead to formation of well-defined complexes consisting of exact molecular stoichiometries (e.g., protein-ligand complexes).
- Non-specific interactions – those due to the hard-core repulsion, electrostatic repulsion and/or attraction, weak hydrophobic attraction, van der Waals potentials, etc. The net effect of these interactions may be repulsive or attractive; while they may lead to the formation of complexes or loosely-bound aggregates, these typically do not exhibit a well-defined stoichiometry or oligomeric state.
CG-MALS is adept at characterizing both specific and non-specific interactions to determine the magnitude of the interaction and the true molecular stoichiometry (when applicable) of resultant complexes and oligomers. While not as sensitive or rigorous, Dynamic Light Scattering (DLS) offers microwell-plate-based, high-throughput screening of interactions and their dependence on buffer conditions.
Biomolecular Interactions (specific)
Ubiquitous in all life forms and central to any research or development program involving biomolecules, the interactions between proteins, nucleic acids, peptides and other biomacromolecules lie at the heart of biology, physiology, and medicine. By providing a direct window into molar mass and size, light scattering offers a unique and powerful approach to analyzing:
- protein-protein or protein-oligonucleotide binding including enzyme-inhibitor or antibody-antigen complexes
- native oligomerization central to the activity of many enzymes
- kinetics of association and self-assembly or dissociation of protein assemblies or amyloid-beta fibrils
CG-MALS excels at analyzing multi-protein assemblies.
MALS is a scientifically rigorous, first-principles technique for determining the weight-average molar mass of macromolecules in solution. As described in Interactions Theory, analysis of molar mass as a function of concentration or composition leads directly and intuitively to the determination of binding affinity (usually as the equilibrium dissociation constant, KD) and absolute molecular stoichiometry.
This analysis is carried out via CG-MALS which combines a composition-gradient preparative device optimized for light scattering measurements, the Calypso™, with a MALS detector such as the DAWN™ and a concentration detector. The CALYPSO™ software not only controls the composition gradient preparation and delivery but acquires and analyzes MALS data in terms of interactions.
The CALYPSO software offers an unparalleled set of association models with the flexibility to describe common as well as unusual complexes, all of which may be found in nature.
Biomolecular Interactions (non-specific)
The cellular environment, at an average macromolecular concentration of 300 – 400 mg/mL, naturally forces proteins, oligonucleotides and other biomacromolecules into close proximity with each other. The short intermolecular distances mean that weak, non-specific interactions may have a significant impact on the behavior of the molecules and modulate their biological function. Changes to specific binding behavior and other functionality arising from this 'molecular crowding effect' are poorly understood since most binding studies take place in purified and dilute solutions. Non-specific protein-protein interactions are also of interest in the formulation of biotherapeutics such as monoclonal antibodies and other biologic drugs.
Light scattering offers the most popular and versatile methods for characterizing non-specific interactions in macromolecules:
- For high-throughput screening of colloidal stability, dynamic light scattering (DLS) has proven to be of great utility. The DynaPro™ Plate Reader measures the mutual diffusion coefficient Dm of biotherapeutic macromolecules to assess colloidal stability via the diffusion interaction parameter kD.
- CG-MALS measures non-specific interactions via the second virial coefficient A2. The Calypso may be programmed to measure A2 automatically as a function of buffer composition, or to measure three values in one method: the A2 of two different macromolecules, and the cross-virial coefficient A11 indicating interactions between the two molecules.
- The two most common factors in non-specific protein-protein interactions are hard-core repulsion and charge-charge repulsion. The magnitude of hard-core repulsion may be calculated from the hydrodynamic radius, measured by DLS, while Electrophoretic Light Scattering (ELS) determines net molecular charge. These values are measured simultaneously in the ZetaStar™. After subtracting the hard-core and charge-charge interaction from the total interaction, the remaining magnitude quantifies the attractive components which lead to colloidal instability.
- Low-volume, simultaneous analysis of A2 and kD are facilitated in multiple Wyatt detectors. The cuvette-based DynaPro ™ NanoStar™ and ZetaStar measure DLS and 90° static light scattering (SLS) in separate, optimized detection channels. The same measurements may be performed in a MALS detector such as the DAWN or miniDAWN™ equipped with a WyattQELS™ DLS module and a microCuvette™ or Microbatch Kit.
- The analysis of interactions at high concentrations can be quite inaccessible and complicated. Thanks to theoretical developments pioneered by Dr. Allen P. Minton1 of the NIH, CG-MALS is one of the very few techniques available for carrying out and analyzing such studies. A variant of this theory is implemented in the CALYPSO software for use in automated or manual concentration gradient MALS experiments.
1 Minton, A. P. Static light scattering from concentrated protein solutions, I: General theory for protein mixtures and application to self-associating proteins. Biophys Journal 2007, 93, 1321–1328.
Resources
Application Notes
Selected References
Attri, A. K.; Fernández, C.; Minton, A. P. Self-association of Zn-insulin at neutral pH: investigation by concentration gradient static and dynamic light scattering. Biophys. Chem. 2010, 148, 23-27.
Folta-Stogniew, E. J. Macromolecular Interactions: Light Scattering. In Encyclopedia of Life Sciences; Wiley: 2001.
Halling, D. B.; Kenrick, S. A.; Riggs, A. F.; Aldrich, R. W. Calcium-dependent stoichiometries of the KCa2.2 (SK) intracellular domain/calmodulin complex in solution. J. Gen. Physiol. 2014, 143, 231-252.
Monterroso, B.; Alfonso, C.; Zorilla, S.; Rivas, G. Combined analytical ultracentrifugation, light scattering and fluorescence spectroscopy studies on the functional associations of the bacterial division FtsZ protein. Methods 2013, 59, 349-362.
Some, D. Light scattering based analysis of biomolecular interactions. Biophys. Rev. 2013, 5, 147-158.
Instrumentation for Interactions
Composition-Gradient System

Calypso™ - Composition-gradient preparative system + software for carrying out and analyzing CG-MALS interaction analyses. Requires a MALS detector.
MALS Detectors

DAWN™ - The most sensitive MALS detector available, anywhere. Incorporates detectors at 18 angles to determine molar masses from 200 Da to 1 GDa and radii from 10 – 500 nm.
- Standard option: ambient temperature
- Heated/cooled option: -15 °C to +150 °C
- High-temperature option: ambient to +210 °C
The DAWN offers special options to handle fluorescent samples: fluorescence-blocking filters and an infrared, 785 nm laser.
miniDAWN™ - Second only to the DAWN in sensitivity. Incorporates detectors at 3 angles to determine molar masses from 200 Da to 10 MDa and radii from 10 – 50 nm. Ambient only.
DynaPro™ NanoStar™ - With sample volumes as small as 1.25 µL and temperature control spanning -15 °C to +150 °C, the NanoStar goes above and beyond traditional cuvette-based DLS instruments: it offers an optimized static light scattering detector (in parallel to the DLS detection system) which may be used with the CALYPSO software to perform CG-MALS experiments with very little sample. Samples must be prepared manually, without benefit of the Calypso's automation.
Dynamic Light Scattering Detectors

The CALYPSO™ software does not analyze DLS data, but can be set to trigger DLS data acquisition by Wyatt’s ASTRA™ software.
WyattQELS™ - A dynamic light scattering (DLS) module which integrates into a DAWN or miniDAWN MALS detector to provide simultaneous DLS measurements in the same scattering volume.
DynaPro™ NanoStar™ - With sample volumes as small as 2 µL and temperature control spanning -10 °C to +120 °C, the NanoStar goes above and beyond traditional cuvette-based DLS instruments: it offers an optimized static light scattering detector (in parallel to the DLS detection system) which may be used with the CALYPSO software to perform CG-MALS experiments with very little sample. The NanoStar does double duty as an online DLS detector by installing its optical collection fiber into a Wyatt MALS detector.
DynaPro™ ZetaStar - The only Wyatt instrument capable of measuring ELS, DLS, and SLS for walk-up or automated measurements of size, particle concentration, and zeta potential. For DLS and SLS measurements, the ZetaStar can measure with as little as 2µL and over a temperature range of -10 °C to + 120 °C. Add the dip cell to the quartz cuvette and measurements of zeta potential with as little as 65 µL of sample. The ZetaStar can also be paired with a Wyatt MALS detectors to act as an online DLS detector by connecting the optical collection fiber.
DynaPro™ Plate Reader - The only commercially available instrument that measures DLS and SLS, for size and molar mass, directly in situ in standard 96, 384 or 1536 microwell plates. The non-perturbing DynaPro Plate Reader offers integration with robot liquid handlers and multi-technique plate-based assay protocols. Measures second virial coefficient A2 and diffusion interaction parameter kD. Temperature-controlled over 4 °C to 85 °C.
Refractive Index Detector

Optilab™ - A unique on-line differential refractometer for measuring concentration of any macromolecule, regardless of chromophores. The high-concentration Optilab accommodates protein concentration up to 180 mg/mL.
Software
CALYPSO™ - The CALYPSO software orchestrates CG-MALS analysis of biomolecular interactions, controlling sample preparation and delivery, data acquisition and data analysis. Provides the most extensive suite of association models available to determine the affinity and absolute molecular stoichiometry of macromolecular interactions including protein-protein binding, binding of aptamers and peptides or proteins, etc., as well as first-order analysis of reaction kinetics.