Are all exosomes essentially alike?
Are all exosomes essentially alike? While proteomic, lipidomic and genomic content, as well as specific function, certainly vary with cell line and environment, common wisdom has held that the vast majority of bionanoparticles produced by cells generally have the same structure and pathway of generation. A tool for differentiating exosomes was needed in order to address this question.
Field-flow fractionation combined with multi-angle and dynamic light scattering (FFF-MALS-DLS) has increasingly received recognition as a key tool in exosome research[1,2]. FFF-MALS-DLS combines true size-based separation using an Eclipse™ FFF system with independent determination of size and structure by a DAWN™ light scattering instrument. It can determine highly accurate size distributions and particle concentrations in native solution.
Perhaps most importantly, the size fractions may be isolated and automatically collected for further off-line investigation using a standard fraction collector.
All exosomes are not alike
Getting back to our question: in a recent breakthrough, H. Zhang et al. in their Nature Cell Biology paper "Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric-flow field-flow fractionation" DOI 10.1038/s41556-018-0040-4 [3] have discovered that all exosomes do not have the same essential nature. Applying FFF-MALS-DLS, they have identified three distinct types of small extracellular vesicles, which they have termed:
- Exo-L, large exosome vesicles (90-120 nm);
- Exo-S, small exosome vesicles (60-80 nm); and
- Exomeres, non-membranous nanoparticles which are the smallest of the three—on average, about 35 nm in diameter.
These three types are grouped by size and other biophysical properties; they are also distinguished from each other by structure (outer membrane or not), protein content, degree of N-glycosylation, metabolite content, DNA/RNA profiles and corresponding function. However, these groupings are conserved across most cell lines, indicating that they are essentially independent classifications of bionanoparticle with correspondingly distinct origins and function. The significance of these findings for cancer research are discussed in Scientists Discover New Nanoparticle, Dubbed Exomeres.
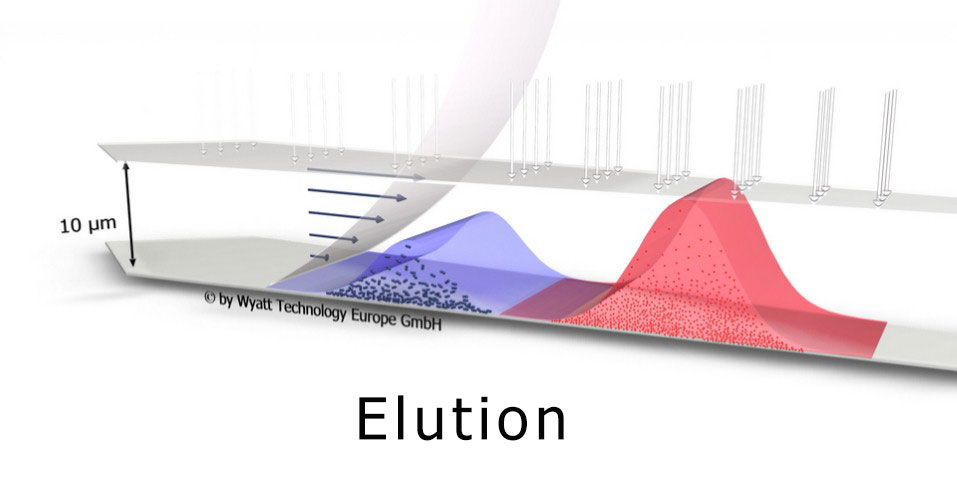
Field-flow fractionation gently separates particles by size from 1 - 1000 nm. The principle of operation balances diffusion with a cross-flow field.
Enabled by FFF-MALS-DLS
The discovery of exomeres is of particular significance, as this class of bionanoparticles had not previously been recognized. Their identification was enabled by the unique features of FFF-MALS-DLS, which separates particles by size prior to quantification by highly sensitive light-scattering instruments. This eliminates some of the pitfalls of non-separating techniques such as nanoparticle tracking analysis (NTA) and standard batch dynamic light scattering (DLS) where the presence of large particles obscures the presence of small particles.
To a large degree the accepted paradigm was a result of characterization and isolation tools that could not really differentiate between sub-types. On the one hand, simplistic size-based characterization tools such as NTA or resistive pulse sensing provided size distributions but nothing more, while sophisticated analyses of protein and nucleotide content produced huge data sets with no clearly defined groupings.
The ability of FFF-MALS-DLS to separate, characterize and isolate the three types of extracellular vesicles extends well beyond traditional means such as cytometry. It was a key enabler for this discovery. The protocol for replicating the Zhang et al study using the Eclipse-DAWN system has been published [5] by Zhang and Lyden of Weill Cornell Medicine in New York City.
Why MALS and DLS?
MALS and DLS both measure nanoparticle size, but are highly complementary:
- MALS determines rms radius from 10 nm to 500 nm, and is about 20x more sensitive than DLS
- DLS determines hydrodynamic radius for 0.5 nm - 200 nm (the upper limit is flow-rate dependent)
- The combination of MALS + DLS is indicative of shape and structure
The DAWN can incorporate an optional DLS module that utilizes the same flow cell and laser, making it versatile indeed. The graphs to the right illustrate how FFF-MALS-DLS provides a complete solution to exosome separation and characterization: MALS takes over where DLS is insufficient.
Beyond FFF-MALS-DLS
FFF-MALS-DLS is a powerful platform for separating and characterizing exosomes and other nanoparticles. In addition to light scattering detectors, it supports additional online detectors such as UV/Vis absorption, fluorescence, differential refractometry with the Optilab™, and even mass spectrometry for extended analyses.
As mentioned earlier, while the Zhang paper represents a breakthrough in exosome research, it is not the first to make use of the Eclipse/DAWN system. In fact, the same setup had previously been employed by Yang et al. [4] to demonstrate that there is a significant difference in size between urinary exosomes from healthy controls and patients with prostate cancer, identifying a potentially life-saving biomarker. Additional publications making use of FFF-MALS-DLS to characterize exosomes have been published [6-8].
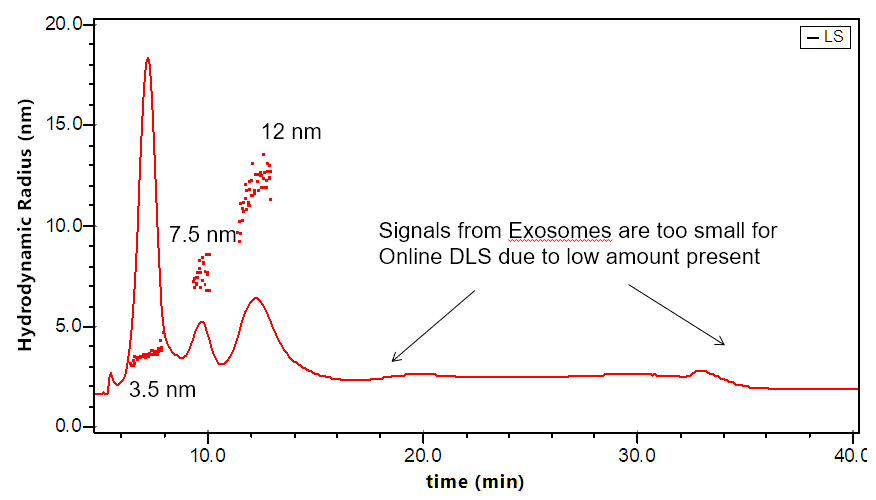
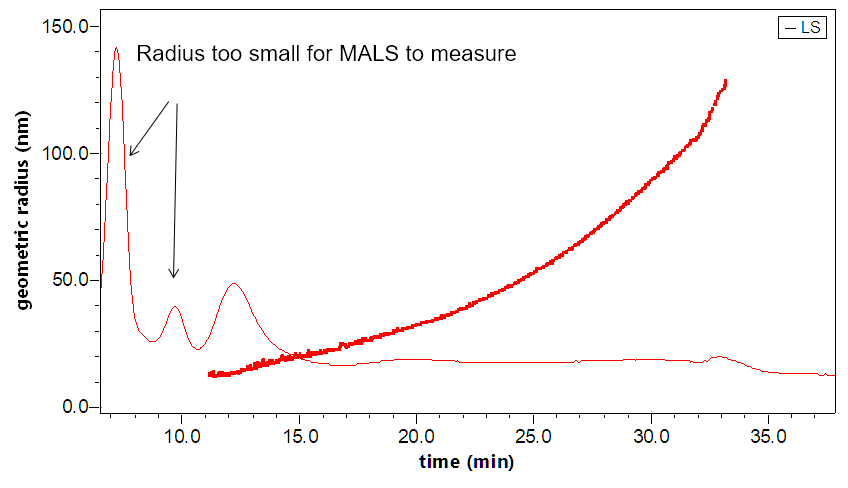
Separation and characterization of exosomes by FFF-MALS-DLS. The triple combination allows for unique characterization and understanding of these bionanoparticles. Utilizing MALS and DLS in tandem extends the range of size measurements, which has proven critical based on recent published results.
Learn more
If you would like to learn more about the capabilities and benefits of FFF-MALS-DLS, please visit our Request Product Info page or contact us at info@wyatt.com
References- Sitar S. et al. (2015) "Size Characterization and Quantification of Exosomes by Asymmetrical-Flow Field-Flow Fractionation", Anal. Chem. 87, pp. 9225-9233 DOI: DOI: 10.1021/acs.analchem.5b01636
- Petersen K.E. et al. (2014) "A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM", Anal. Bioanal. Chem. 406, pp. 7855-7866 DOI: 10.1007/s00216-014-8040-0
- Zhang H. et al. (2018) "Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation", Nat. Cell Bio. 20, pp. 332–343 DOI: 10.1038/s41556-018-0040-4
- Yang J.S. et al. (2017) "Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry", Anal. Chem. 89, pp. 2488-2496 DOI: 10.1021/acs.analchem.6b04634
- Zhang H. and Lyden D. (2018) "A protocol for Asymmetric-Flow Field-Flow Fractionation (AF4) of small extracellular vesicles", Protocol Exchange, 19 February 2018, DOI: 10.1038/protex.2018.002
- Agarwal K. et al. (2015) "Analysis of Exosome Release as a Cellular Response to MAPK Pathway Inhibition", Langmuir 31(19), pp. 5440-5448 DOI: 10.1021/acs.langmuir.5b00095
- Ashames A. (2015) "Development of separation methods to produce uniform exosomes subpopulations using field-flow fractionation techniques", Ph.D. thesis. Colorado School of Mines DOI: http://hdl.handle.net/11124/17052
- Kang D. et al. (2008) "Proteomic Analysis of Exosomes from Human Neural Stem Cells by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry", J. Proteome Res. 7 pp. 3475-3480 DOI: 10.1021/pr800225z

The Eclipse system combines standard HPLC components with an AF4 controller and channel to separate particles by size, then determines detailed size, shape, molar mass and composition distributions using online multi-angle and dynamic light scattering with additional online detection modes.
