Next steps in biophysical characterization by MALS: ion-exchange and reversed-phase chromatography
Thoroughly explored SEC-MALS, and ready to take the next steps to further enhance your biomolecular characterization capabilities?
SEC-MALS for proteins
For the biophysical characterization community, multi-angle light scattering (MALS) has been closely associated with size-exclusion chromatography (SEC) for well over two decades. The key benefits of SEC-MALS for protein characterization were expounded in a seminal paper by Wen, Arikawa and Philo [1], built on a strong technical foundation of instrumentation and software from Wyatt Technology™. Ever since, SEC-MALS has been an essential analytical technique in biophysical labs, whether in academic or biopharmaceutical settings.
In a typical SEC separation, different protein species—for example, monomers, oligomers, complexes, fragments or simply different proteins—elute at different times and can be evaluated immediately by MALS analysis. The analysis combines signals from a DAWN™, miniDAWN™ or µDAWN™ MALS detector and one or two concentration detectors, usually an Optilab™ differential refractive index (dRI) and/or the HPLC’s UV detector. The primary applications of SEC-MALS are:
- absolute determination of molar mass—independently of elution time, column calibration standards, molecular conformation and non-ideal column interactions;
- characterization of conjugated proteins—glycoproteins, pegylated proteins or surfactant-bound membrane proteins—which do not correspond to globular calibration standards;
- determination of the stoichiometry of protein-protein and protein-nucleic acid complexes as well as native oligomers and aggregates.
Isocratic vs. gradient
Coupling MALS to SEC is an obvious choice. First, SEC (usually) separates by hydrodynamic size, which is (usually) closely related to molar mass, the primary objective of MALS analysis. Second, dRI analysis is straightforward in an isocratic SEC separation, whereas gradient separations tend to produce large changes in refractive index—much, much larger than the change measured at the analyte peak—which can overwhelm and even saturate the dRI detector.
Despite its many advantages for MALS analysis, SEC is subject to several limitations:
- separation of aggregates beyond trimers suffers from low resolution
- analytes with similar size cannot be separated from each other (e.g. protein-filled vs. empty detergent micelles, antibody-drug conjugates with different drug-antibody ratios, or similar-sized proteins)
- sample load is limited and peaks are broad, rendering poor signal-to-noise in the analysis of very-low-molecular-weight samples such as peptides and small proteins
- successful method development can be a tedious and lengthy process and limited in the number of parameters available for adjustment
Other types of chromatography, such as ion-exchange (IEX) and reversed-phase (RP), separate by parameters other than size, offer more robust and tunable separation than SEC and can overcome these limitations. So why don’t we hear more about their use with MALS?
Due to the mobile-phase gradient and the operation of the proportioning valve that induces composition noise, dRI is not a good choice of concentration detector for these techniques. However, MALS plus a UV/Vis absorption detector can most certainly be used with IEX, RP and other types of chromatography, with one main caveat: you need to know the UV extinction coefficient with reasonable accuracy, and for that you need to know which protein is eluting in each peak.
MALS without dRI
In many characterization tasks, this is not an issue at all. You know which protein is eluting; extinction coefficients can be calculated from the sequence or measured separately using ASTRA™'s Extinction coefficient from RI peak method using SEC; variants and aggregates have (to within experimental uncertainty) the same extinction coefficient as monomers. Even IgG fragments tend to have about the same extinction coefficient as the monomer. Extinction coefficients at wavelengths other than 280 nm, e.g. the peptide backbone absorption at 214 nm, may be less variable than at 280 nm. In these cases, IEX-MALS or RPC-MALS offer opportunities to enhance separation, resolving and characterizing species that are challenging on SEC.
On the other hand, MALS analysis with concentration from dRI offers some very powerful benefits: universal protein response to identify unknown protein species, and the ability to couple with UV for conjugate analysis. In a mild refractive-index gradient such as employed in IEX, there may be a way to incorporate dRI (more on this below), but to the best of our current understanding it cannot be used with more extreme gradients such as are used in RPC or HIC. With such disparate mobile phase refractive indices, Imperfect gradient mixing produces dRI noise which is just too high for proper MALS calculations. In the future, better mixers may lead to acceptable dRI noise in gradient HPLC.
IEX-MALS and RPC-MALS sound interesting, but do they really work? The proof is in the peer-reviewed publication pudding, as they say. Coupling of light scattering to RP, IEX and hydrophobic interaction chromatography was successfully demonstrated long ago [2-5] and more recently [6], so they definitely work and produce viable results. Figure 3 demonstrates robust RPC-MALS analysis performed in-house at Wyatt’s Applications Lab over 20 years ago. It’s likely that at the time, there was still so much to progress to be made with SEC-MALS alone that the other techniques were perennially relegated to “for later review”.
IEX-MALS makes a comeback
Now it seems that “later” has arrived [7]: in a just-released article titled “Coupling Multi Angle Light Scattering to Ion Exchange chromatography (IEX-MALS) for protein characterization” by Amartely et al., freely available online at www.nature.com/articles/s41598-018-25246-6, the authors make the arguments for these IEX-MALS benefits:
- oligomers are well-resolved including (at least) dimer-tetramer-hexamer-octamer
- aggregates elute after the monomer, minimizing contamination of the monomer signal
- large quantities of peptides or small proteins can be loaded onto the column to improve signal-to-noise ratio and, ultimately, molar mass measurement—especially important when the sample is aggregation-prone and cannot be concentrated
- a full range of parameters including salt and pH gradients can be tweaked to resolve molecules with similar size that co-elute in SEC, but have different sequences, glycosylation patterns or other modifications
Other types of chromatography, such as ion-exchange (IEX) and reversed-phase (RP), separate by parameters other than size, offer more robust and tunable separation than SEC and can overcome these limitations. So why don’t we hear more about their use with MALS?
Explore more
Have you have encountered difficulties in SEC-MALS analysis pertaining to the separation method? Would you like to explore the use of IEX-MALS, RPC-MALS or other chromatographic variants? If so, we invite you to contact us atsupport@wyatt.com. Our Application Scientists are eager to find out about your needs and help solve your MALS application challenges, even if that means going outside of our SEC comfort zone. The results may surprise all of us!
References
- J. Wen, T. Arakawa, J. S. Philo. (1996) “Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions” Anal. Biochem. 240 pp. 155-166.
- R. Mahtre and I. S. Krull (1993) “Determination of on-line differential refractive index and molecular weight via gradient HPLC interfaced with low-angle laser light scattering, ultraviolet and refractive index detection” Anal. Chem. 65 pp. 283-286.
- R.M. Mahtre and I.S. Krull (1992) “Interfacing gradient elution ion-exchange chromatography and low-angle laser light-scattering photometry for analysis of proteins” J. Chromat. 591 pp. 139-148.
- I.V. Astafieva, G.A. Eberlein and Y.J. Wang (1996) “Absolute on-line molecular mass analysis of basic fibroblast growth factor and its multimers by reversed-phase liquid chromatography with multi-angle laser light scattering detection” J. Chromat. A 740 pp. 215-229.
- S. Iuliano, J. R. Fischer, M. Chen and W. J. Kelly (2002) "Rapid analysis of a plasmid by hydrophobic-interaction chromatography with a non-porous resin: J. Chromat. A 972(1), pp. 77-86.
- M. Onsberg et al. (2013) “Light scattering coupled with reversed phase chromatography to study protein self-association under separating conditions” J. Chromat. B 938 pp. 60-64.
- H. Amartely et al (2018) “Coupling Multi Angle Light Scattering to Ion Exchange chromatography (IEX-MALS) for protein characterization”, Sci. Rep. accessed online at www.nature.com/articles/s41598-018-25246-6 May 2, 2018.

Figure 1. A SEC-MALS system for protein analysis includes HPLC with SEC column and UV detector, a MALS instrument (DAWN, miniDAWN or μDAWN) and dRI detector (Optilab), and ASTRA® software.
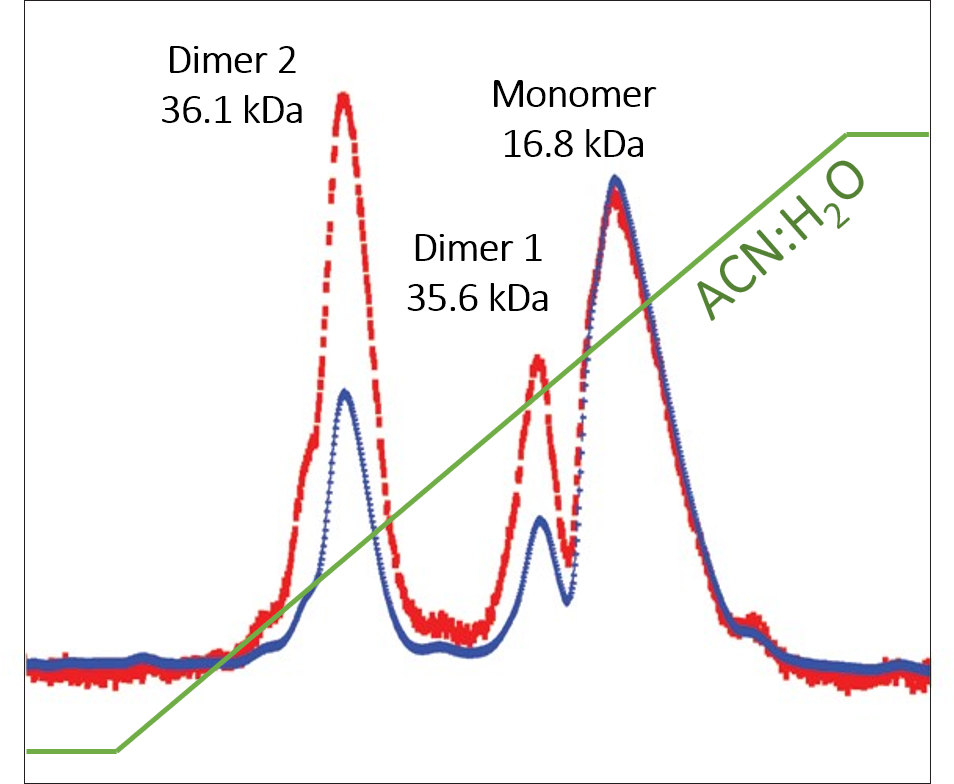
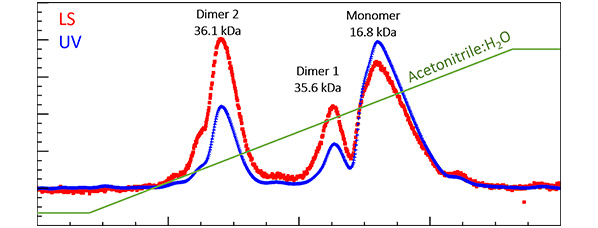
Figure 2. Under reversed-phase separation, fibroblast growth factor is found to contain three species. MALS with UV indicates that two are dimers, with equivalent molar masses but quite different hydrophobicity. The differences in hydrophobicity may arise from conformational differences [4].
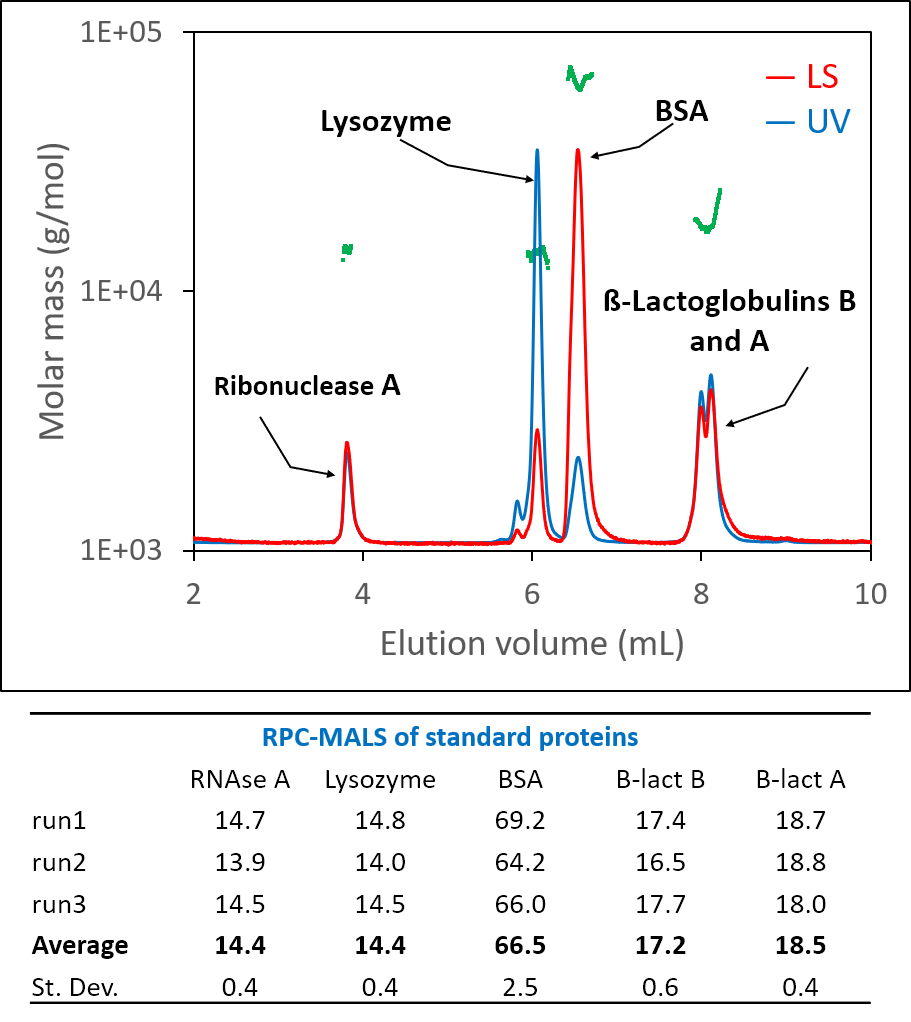
Figure 3. Standard proteins analyzed by RPC-MALS-UV.
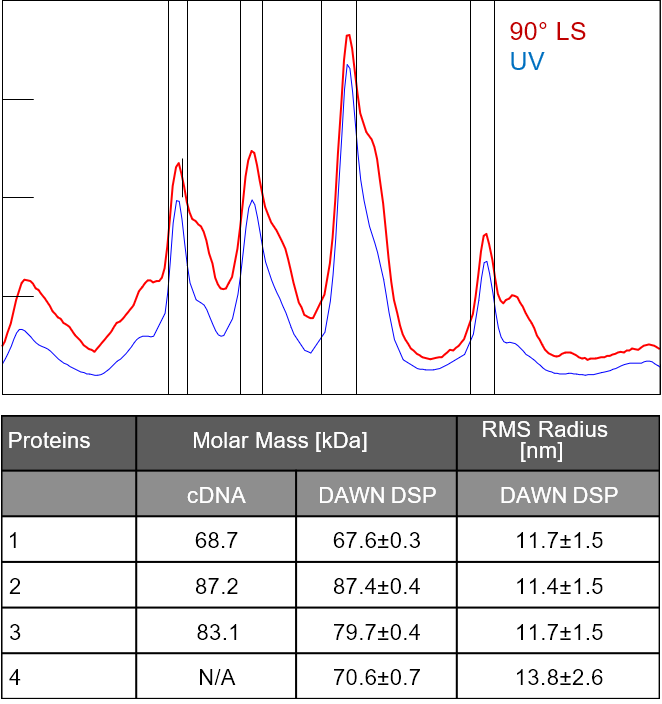
Figure 4. Wheat gluten analyzed by RPC-MALS. See application note Wheat Protein Characterized by RPC-MALS-AN8001
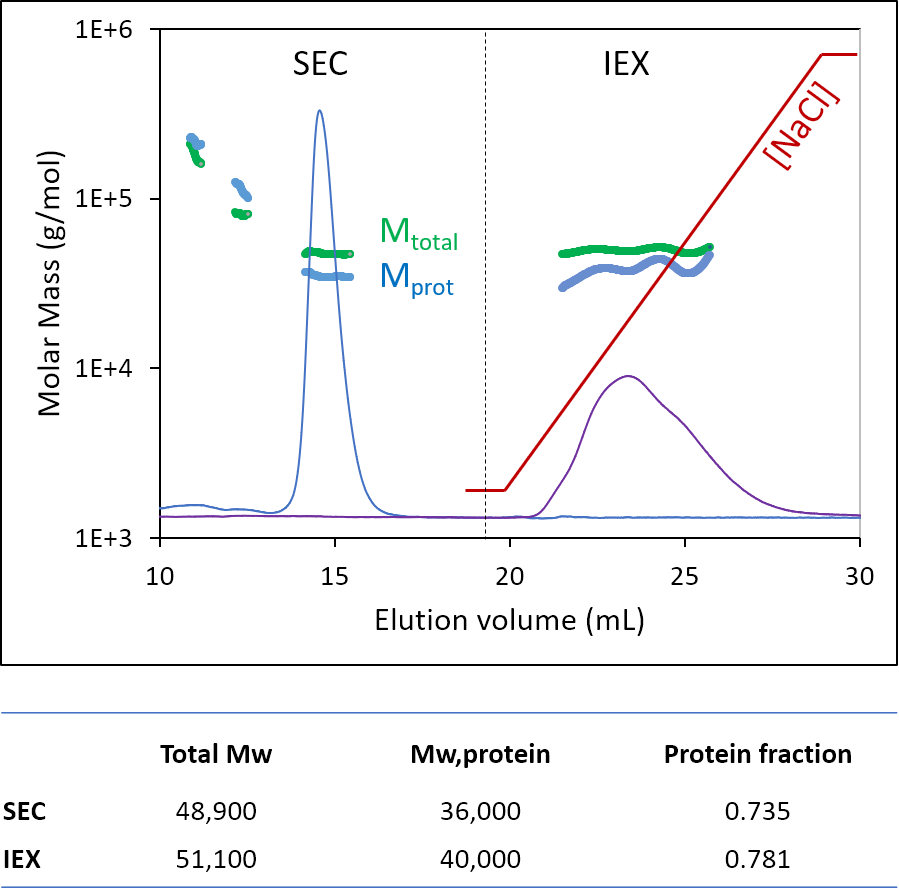
Figure 5. A glycoprotein analyzed by SEC-MALS and IEX-MALS using ASTRA's Protein Conjugate Analysis. In this initial demonstration the IEX-MALS analysis is not as robust as the SEC-MALS analysis, but overall results are quite similar. Data courtesy Hadar Amartely and Mario Lebendiker, Hebrew University of Jerusalem, Israel.
