PIPPI: developing a new toolkit for predicting protein aggregation

The problem
Current approaches for predicting long-term storage stability of protein therapeutics suffer from empiricism when extrapolating across accelerated stress conditions such as increased temperature or agitation. The kinetics of the formation of aggregates ranging from dimers to large protein particles are not adequately understood.
The challenge
Aggregation involves multiple pathways. Rate controlling steps involve partially folded intermediates that occur at low relative populations and are difficult to isolate and study. From these intermediates evolve a full spectrum of protein aggregate sizes. Both the intermediates and final size spectrum need to be studied over time to understand individual kinetics.
The goal
Develop a toolkit for predicting aggregation, with a focus on small numbers of partially folded intermediates and the full spectrum of aggregate sizes.
These were the problem, challenge and goal assigned to Lorenzo Gentiluomo when he accepted the PIPPI fellowship. PIPPI, which stands for Protein-excipient Interactions and Protein-Protein Interactions, is an academic-industrial consortium funded by the European Horizon 2020 program. It is an Innovative Training Network (ITN) addressing the challenges in formulation of protein-based drugs. PIPPI’s mandate is to combine systematic investigations of the physico-chemical properties of a number of proteins with an in-depth understanding of the molecular interactions behind their macroscopic behavior. Its overall objective is to develop methodologies, tools and databases to guide the rational formulation of robust protein-based therapeutics.
Lorenzo is a Ph.D. student supervised by Prof. Dr. Wolfgang Friess of the Ludwig Maximilians University (LMU) Dept. of Pharmacy, Munich. He carries out much of his work under Dr. Dierk Roessner at Wyatt Technology™ Europe GmbH (WTE) in Dernbach, Germany. WTE is an eager partner in the PIPPI consortium, contributing lab and office space, as well as the use of Wyatt’s unique instrumentation needed for understanding protein aggregation pathways. Having studied chemistry at La Sapienza University of Rome and the University of Jyväskylä, culminating in a M.Sc. cum laude in organic and biomolecular chemistry, while simultaneously working as formulation scientist at Henkel and as a young researcher at Jyväskylä University, Lorenzo is well-prepared to take on the challenge.
- Automating methods for quantifying aggregation kinetics by high-throughput static and dynamic light scattering (HT-DLS) and special size-exclusion chromatography-multi angle light scattering (SEC-MALS) setups with modified sampling systems.
- Providing the first systematic studies of aggregation kinetics for an extensive database of proteins formulated in a range of conditions.
- Developing predictive models that integrate knowledge of individual steps in aggregation pathways, such as the initial unfolding step, the reversible association, and the interactions between partially folded proteins.
- Exploiting information-rich techniques such as asymmetric-flow field-flow fractionation (AF4) or raster image correlation spectroscopy, in order to provide additional details about the time evolution of monomer and oligomer species and so refine aggregation models.
As of the date of this writing (July 16, 2018) Lorenzo has spent two years in the program, during which time he has learned the ins and outs of SEC-MALS, µSEC-MALS , CG-MALS, HT-DLS and differential viscometry. The tools he has used include
As of the date of this writing (July 16, 2018) Lorenzo has spent two years in the program, during which time he has learned the ins and outs of SEC-MALS, µSEC-MALS , CG-MALS, HT-DLS and differential viscometry. The tools he has used include
-
- µDAWN™ plus Optilab UT-rEX with Waters™ ACQUITY™ UPLC™ for fast, low-volume µSEC-MALS analyses;
-
- DynaPro™ Plate Reader for HT-DLS/SLS;
-
- Calypso™ plus DAWN™ for characterization of interactions by CG-MALS;
-
- Eclipse™ DualTec™ for advanced separations of large and small aggregates by AF4;
-
- DynaPro Nanostar™ for quasi-equilibrium studies by DLS and SLS;
-
- Mobius™ for electrophoretic mobility measurements to determine protein charge;
- and last but not least, the Viscostar™ for a novel application of differential viscometry in biotechnology.
The accomplishments
During the first two years of his work, Lorenzo completed the first screening of a series of protein provided by Roche, MedImmune and Novozyme, assessing conformational and colloidal stability across many different formulations. This activity was a key initial step in building up the PIPPI database, and the work done by Lorenzo with WTE’s support constituted about 80% of the total data set to be used by the 15 PIPPI fellows. The database will provide a solution to the current lack of high-value knowledge regarding the protein formulation conditions that result in stable drug products. He will be first author on the manuscript reporting these results.
Using this dataset, he trained an artificial neural network to predict colloidal and conformational stability of mAbs solely on the basis of amino acid composition. This approach could optimize the selection of candidate proteins with the highest potential for successful product development, even before their expression, harvesting and purification, by predicted their melting temperature Tm, aggregation onset temperature Tagg, diffusion interaction parameter kD and monomer loss over thermal stress. Furthermore, the neural network model is expected to reduce the number of formulation conditions (i.e. pH and NaCl concentration) that need to be physically tested during early formulation development. This work has been submitted for publication.
Detailed studies of native/reversible aggregates are rare, and most of the aggregation kinetic literature focuses on non-native/irreversible aggregation. In parallel to the work mentioned above, Lorenzo completed a study of the reversible, native aggregation of one protein, using a series of methods ranging from static and dynamic light scattering to analytical ultracentrifugation and small angle x-ray scattering. This work will also be submitted for publication.
Sounds like significant accomplishments for just two years, but there is more!
- In collaboration with PIPPI fellow Hristo Svilenov and Prof. Gerhard Winter’s group at LMU, he studied the physical stability of monoclonal antibodies by denaturant dilution, using the DynaPro Plate Reader. The work is currently under review and will be published in J. Pharm. Sci.
- In collaboration with PIPPI fellow Aisling Roche and Prof. Robin Curtis of the University of Manchester, he developed a method to predict protein viscosity at high concentration using differential viscometry and the ViscoStar.
- He is collecting additional data to be used in the prediction of the second viral coefficient, exploiting the high-throughput B22 screening capabilities of the DynaPro Plate Reader III to generate complete datasets across many proteins and formulations with just a few µL per sample and within few days. These data will be used by PIPPI fellow Marco Polimeni and Dr. Mikael Lund of Lund University to fit Monte Carlo simulations for the a priori prediction of B22 of small proteins based on amino acid composition.
- In collaboration with PIPPI fellow Sujata Mahapatra, Dr. Werner Streicher of Novozyme, and Prof. Pernille Harris and Prof. Peter Günther of the Technical University of Denmark, he is using CG-MALS to correlate B22 to the structure factor at angle 0° obtain by SAXS to study protein-protein interactions.
- In collaboration with PIPPI fellow Christin Pohl, Dr. Allan Nørgaard of Novozyme, and Prof. Pernille Harris and Prof. Peter Günther of the Technical University of Denmark he is helping to study peptide gel formation by the use of AF4.
- In collaboration with Dr. Christoph Johann of WTE as well as Christin Pohl, Sujata Mahapatra, Dr. Allan Nørgaard, Dr. Werner Streicher, Prof. Pernille Harris and Prof. Peter Günther, he is studying the relative bias between SEC, AF4 and AUC aggregation analyses for a series of proteins.
- In collaboration with PIPPI fellow Maria Laura Greco and Dr. Christopher van der Walle of Medimmune, he is characterizing an antibody-drug conjugate.
The outlook
In the last year of his fellowship, Lorenzo will focus on the screening of stressed formulations to better understand the effect of excipients on protein aggregation. He will use a new high-throughput approach to study protein aggregation, and a machine learning algorithm to create models capable of predicting protein aggregation. Moreover, he foresees developing some new applications for the Wyatt instruments. Lastly—thanks to the PIPPI network—a series of projects in collaboration with the other fellows are in the works. We all look forward to the final publication of each of these exciting studies.
Wyatt Technology welcomes support for, and interaction with, the PIPPI consortium, as part of its company mission, to “… delight its customers by providing outstanding analytical tools, as well as unparalleled levels of personal service, to support life-enhancing macromolecular and nanoparticle science.” The insights produced by Lorenzo’s research will benefit humanity by enabling the development of life-saving medications that are more available, efficacious and lower-cost with fewer side-effects than traditional drugs. As part of that effort, his work will also inform Wyatt’s development of the instruments and software needed by the biopharmaceutical industry to achieve these worthy goals. We look forward to more fruitful interactions with Lorenzo and his colleagues in the coming years!

The PIPPI consortium members and fellows, gathered at MedImmune in Cambridge, UK.
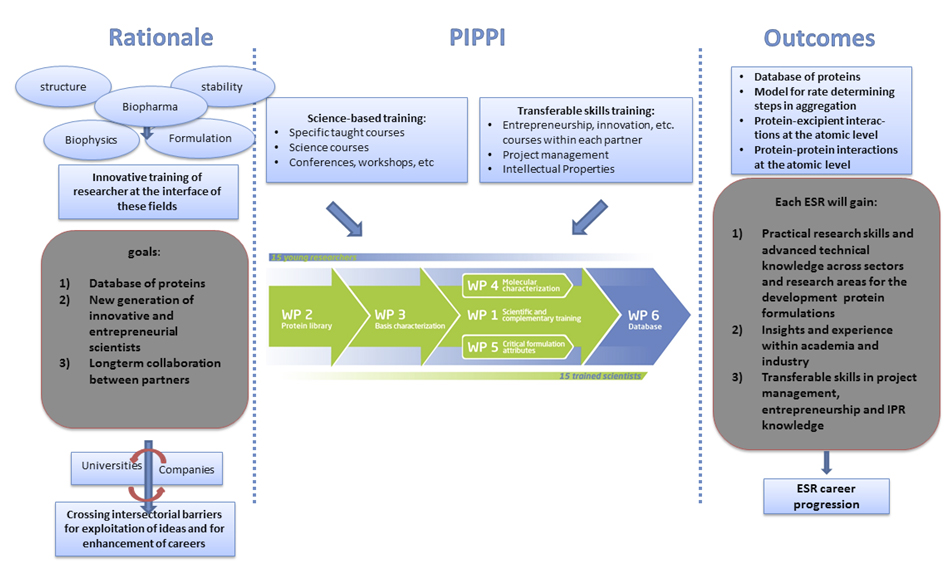
Overview of the PIPPI research and training program. (click to view larger)

PIPPI Fellow Lorenzo Gentiluomo, hard at work in the lab!”

Explaining his research to attendees of Wyatt’s workshop on protein characterization in Dernbach.
“Including Wyatt Technology in the consortium has been instrumental for the success of the project. The access to best-in-class Wyatt instruments and their scientific expertise has been really important to be able to characterize all the proteins in an efficient way. Also the possibility for the PhD students to go to Wyatt and be part of the highly specialized, industrial environment has made it possible for the students to gain knowledge they would otherwise not have obtained.”
