
Obtain Absolute Measurements
Our DAWN® multi-angle light scattering (MALS) instruments allow you to determine absolute molecular weights and sizes without relying on so-called standards, or measurements made in someone else’s lab. Wyatt instruments measure all of the quantities required for determining absolute molar masses directly.

DAWN®
The most sensitive MALS detector available, anywhere. Incorporates detectors at 18 angles to determine molar masses from 200 Da to 1 GDa and radii from 10 – 500 nm.
Don't Assume
While there are many ways to determine the molar masses, or molecular weights, of macromolecules such as proteins, biopolymers and synthetic polymers, only one determines absolute molecular weights in solution, rapidly and effectively: Multi-Angle static Light Scattering (MALS). MALS combines the precise measurements of light scattering intensity and concentrations for a rigorous, first-principles calculation of solution-property molecular weights—making no assumptions of conformation or shape, and no perturbations to the sample.
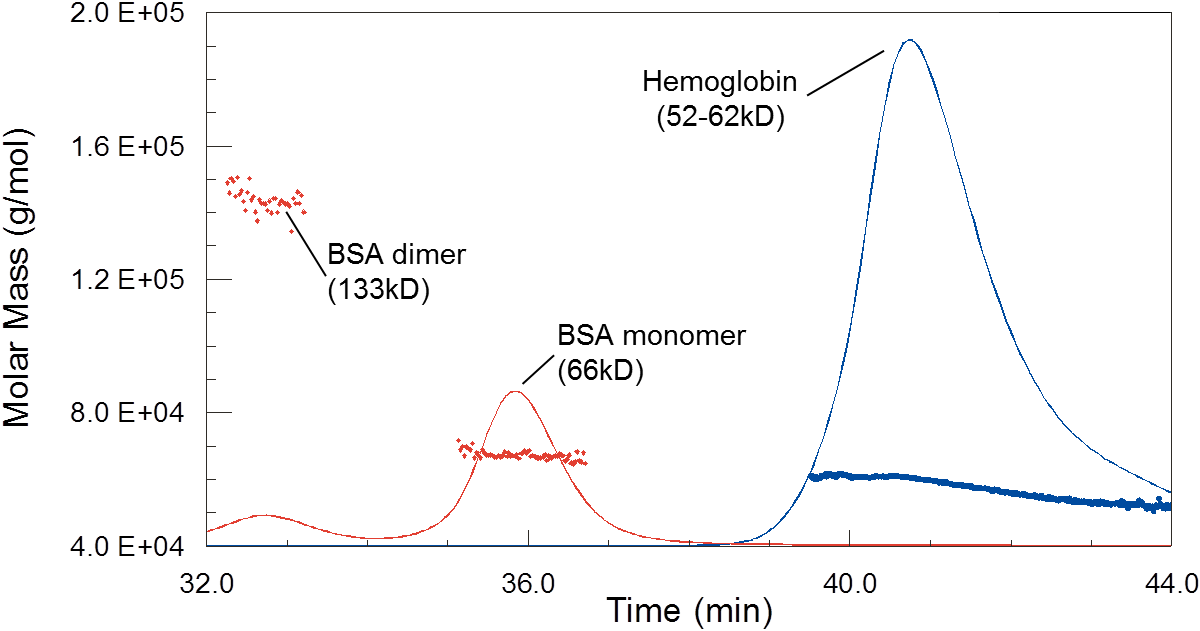
While column calibration would suggest that hemoglobin is much smaller than BSA, MALS shows that they do in fact have similar molar masses. The polydispersity of hemoglobin, seen here as a trailing shoulder on the peak with decreasing molecular weight, is due to dynamic equilibrium between monomers, dimers and tetramers.
Identification
Molar mass is the key to identifying proteins, their oligomers or complexes, yet all too many protein researchers rely on simplistic, invalid analysis of molecular weight by SDS-PAGE or traditional size exclusion chromatography (SEC). These techniques invoke assumptions of conformation and ideal matrix interactions that may fool scientists with ambiguous or completely incorrect results, leading to fundamentally inaccurate interpretation of their data for scientific publications.
Multi-angle light scattering coupled with SEC (SEC-MALS) provides accurate molecular weight determination of proteins, oligomers and complexes, regardless of conformation or non-ideal column interactions. That's because SEC-MALS constitutes a rigorous, first-principles analysis of molar mass that does not rely on retention time or calibration with reference molecules. The only function of the SEC column is to separate molecules by size, while MALS determines molar mass of eluting proteins independently.
Conjugation
Many interesting and useful macromolecules consist of two chemically and physically distinct species, conjugated covalently. This can make it quite difficult to characterize the essential properties of molar mass and size by size-exclusion chromatography since no calibration standards exist to represent the elution behavior of these composites. Often one or both of the primary constituents is not amenable to mass spectrometry.
The combination of multi-angle light scattering (MALS), UV and refractive index (RI) detection with size exclusion chromatography, plus ASTRA®'s Conjugate Analysis algorithm, offers the solution to the characterization of conjugated macromolecules such as:
- Block co-polymers
- Glycoproteins, lipoproteins and nanodiscs
- PEGylated proteins
- Membrane proteins solubilized in detergent micelles
- Protein-polysaccharide vaccines
- Antibody-drug conjugates
Conceptually, a complex consisting of a protein (containing a UV chromophore) and a modifier such as poly-ethylene glycol, or PEG (which does not), can be analyzed by combining information from two distinct measurements. UV absorption determines the protein concentration, while RI determines total macromolecular concentration. The modifier concentration is just the difference between these two (actually, an accurate calculation accounts for the extinction coefficients and differential refractive indices of both species). These two measurements are sufficient to determine the ratio of the protein and modifier in the complex. However, this analysis cannot determine the total mass or oligomeric state of the species.
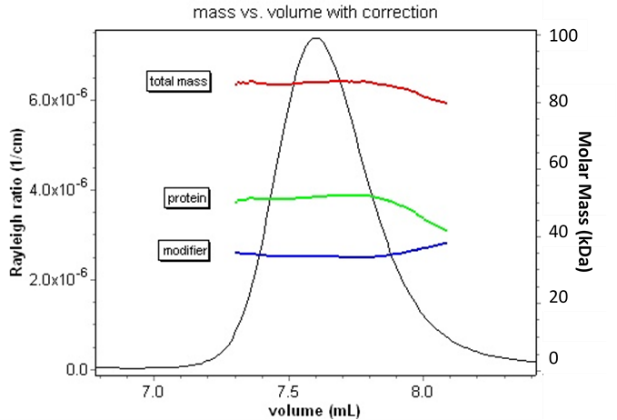
Combining MALS-UV-RI analysis with size exclusion chromatography enables the analysis of conjugated proteins.
The addition of a MALS detector provides the answer to full conjugate characterization. Since light scattering is proportional to the product of molar mass and concentration, the combination of these three signals is sufficient to determine not only the ratio but the actual molar mass, and hence oligomeric state, of each constituent. This information is critical in assessing aggregation of conjugates, measuring the molar mass of block co-polymers, or deciding whether or not a detergent-solubilized membrane protein is present as a native oligomer.
Membrane Proteins
Membrane proteins solubilized with detergent are particularly difficult to analyze by traditional techniques or even by mass spectroscopy because of the surfactant micelle surrounding the protein. Denaturing SDS-PAGE dissociates native oligomers and precludes their identification, while cross-linked mass spectroscopy can create oligomers that do not exist in solution.
Likewise, heavily glycosylated proteins cannot be represented by reference standards or common models for globular proteins, and so are not amenable to analysis by traditional techniques.
These challenges are met by multi-angle light scattering coupled with SEC (SEC-MALS) which can distinguish between a protein and its associated detergent or carbohydrate by combining data from three detectors downstream of SEC separation: UV, MALS and differential refractive index (dRI). ASTRA's Conjugate Analysis algorithm calculates the molar masses of both the proteinaceous component and the conjugated or micellar component. The true oligomeric or complexated state of the protein, as well as the degree of glycosylation, are determined unambiguously.
